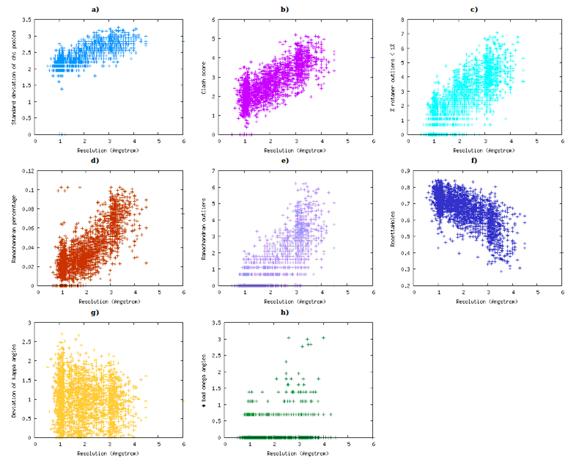
IDP Force Fields Applied to Model PPII-Rich 33-mer Gliadin Peptides | The Journal of Physical Chemistry B

Secondary Structure and Conformational Stability of the Antigen Residues Making Contact with Antibodies | The Journal of Physical Chemistry B
Significant Differences in Physicochemical Properties of Human Immunoglobulin Kappa and Lambda CDR3 Regions
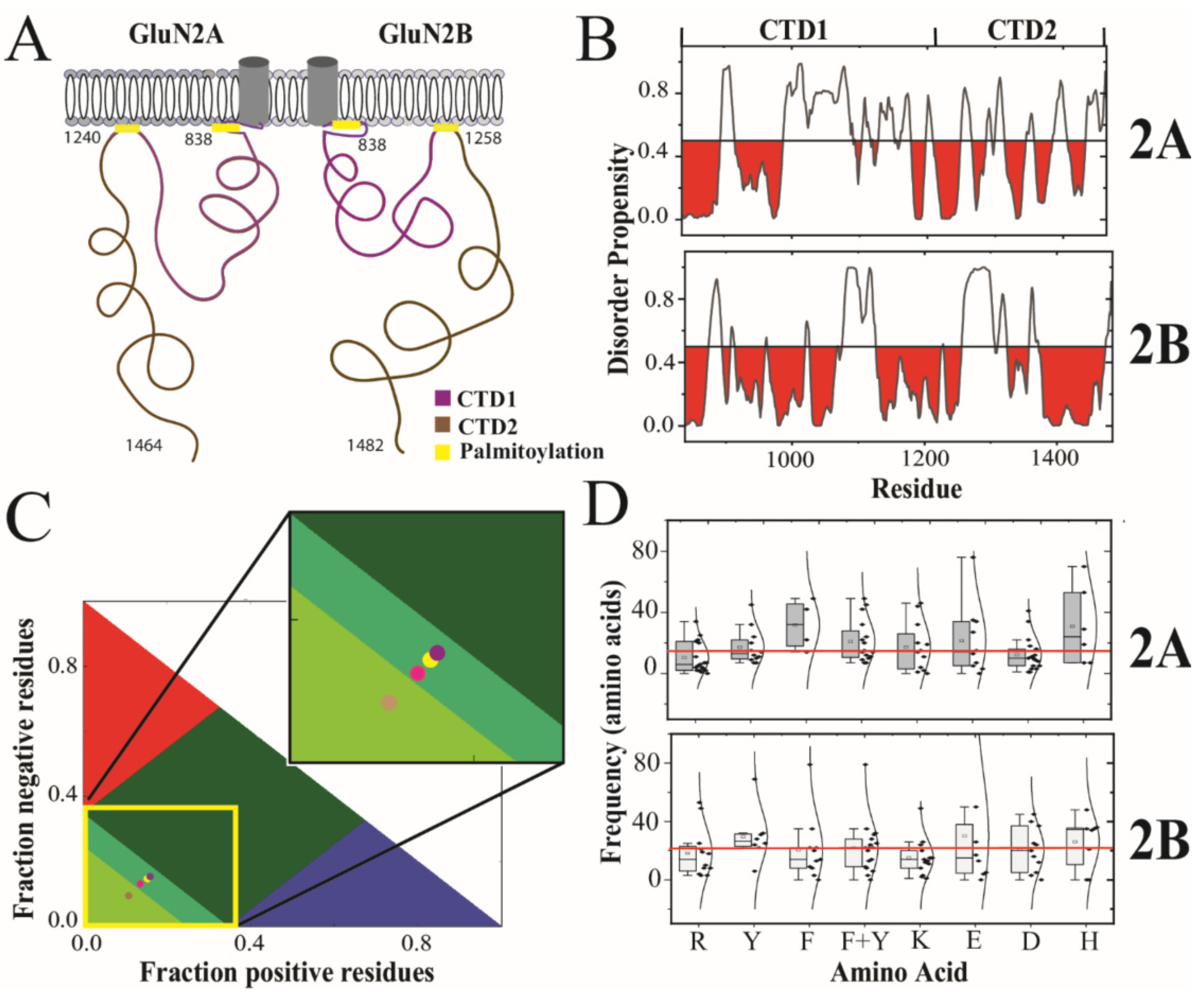
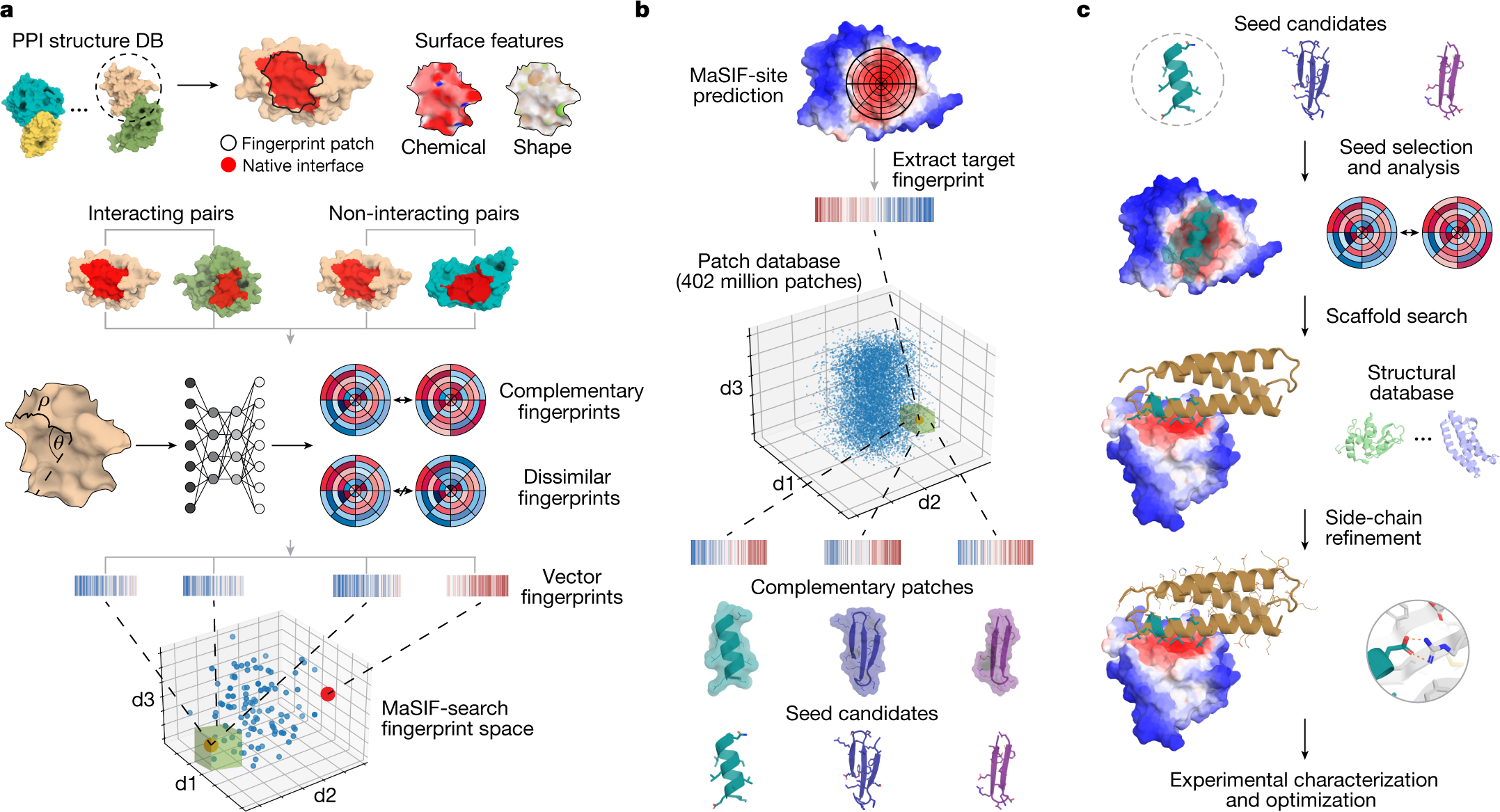
Biomolecules | Free Full-Text | Different Forms of Disorder in NMDA-Sensitive Glutamate Receptor Cytoplasmic Domains Are Associated with Differences in Condensate Formation
Significant Differences in Physicochemical Properties of Human Immunoglobulin Kappa and Lambda CDR3 Regions

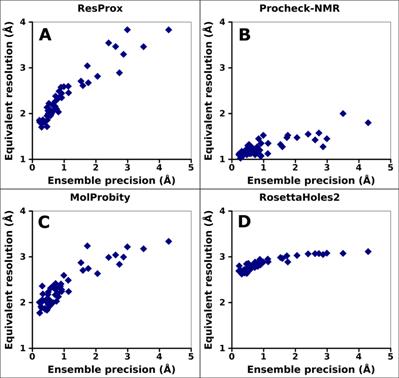
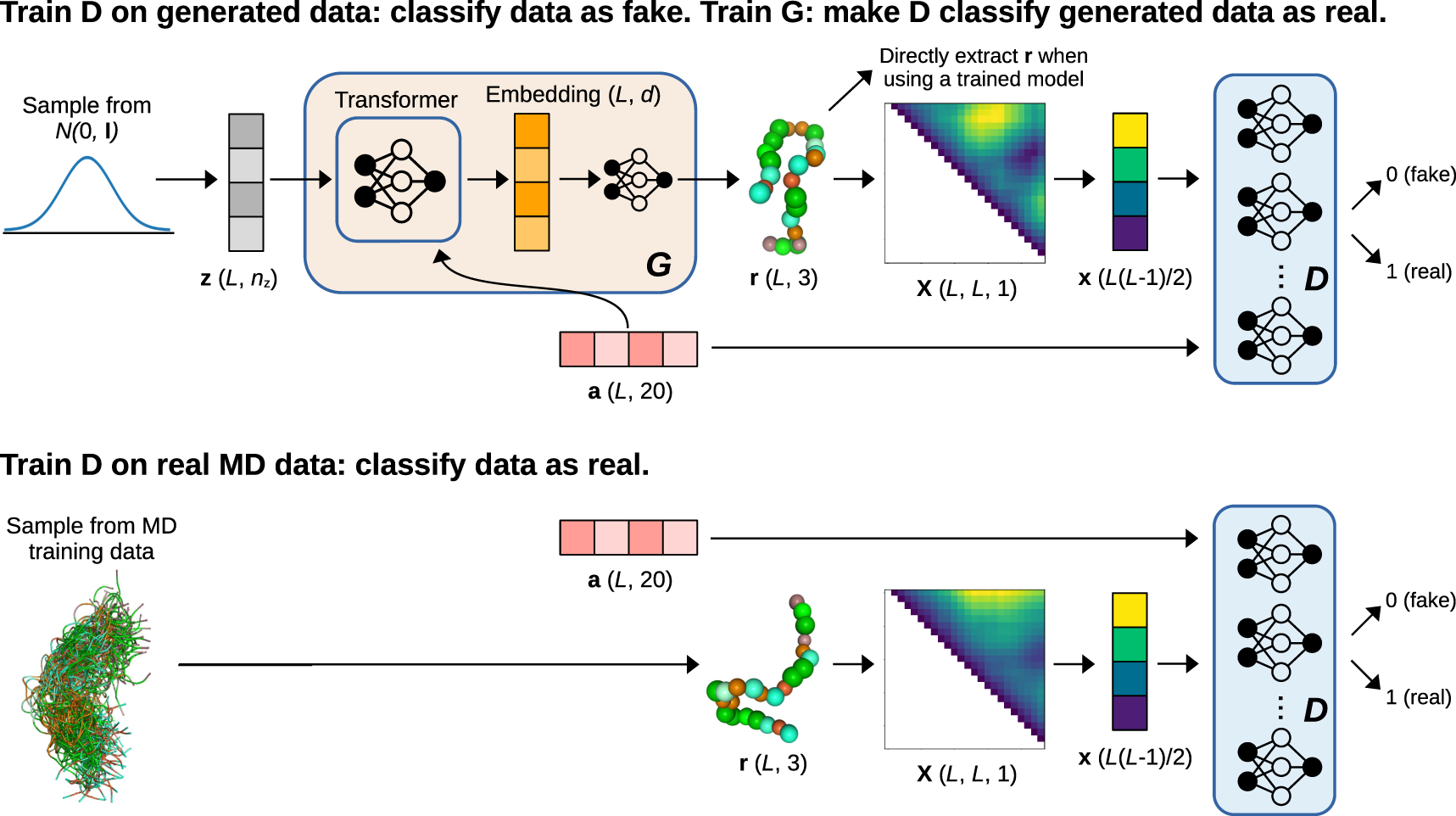
Generation of the configurational ensemble of an intrinsically disordered protein from unbiased molecular dynamics simulation | PNAS

Kink Characterization and Modeling in Transmembrane Protein Structures | Journal of Chemical Information and Modeling
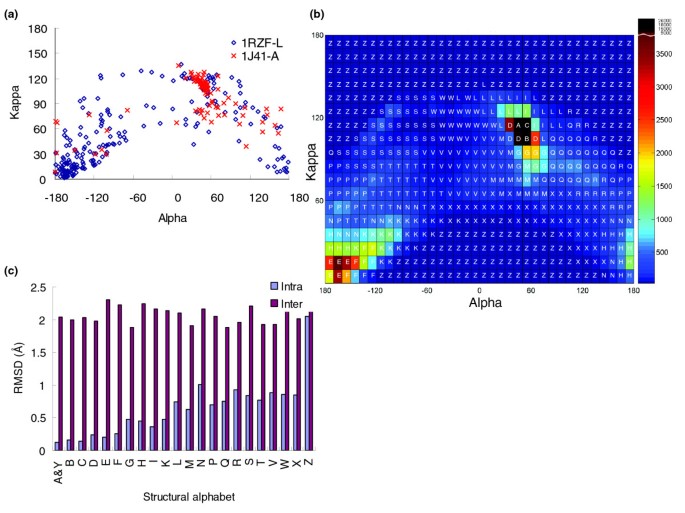
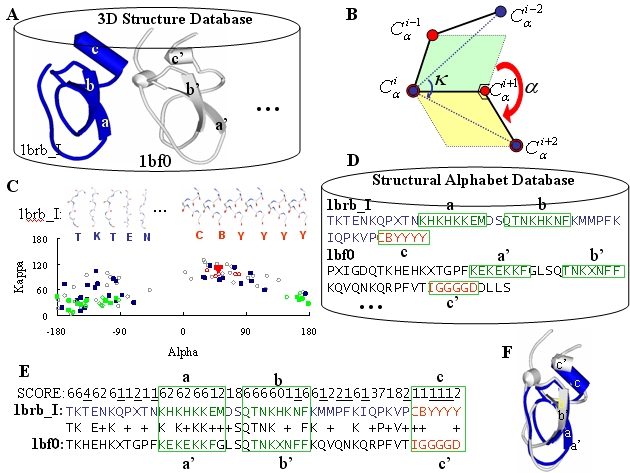
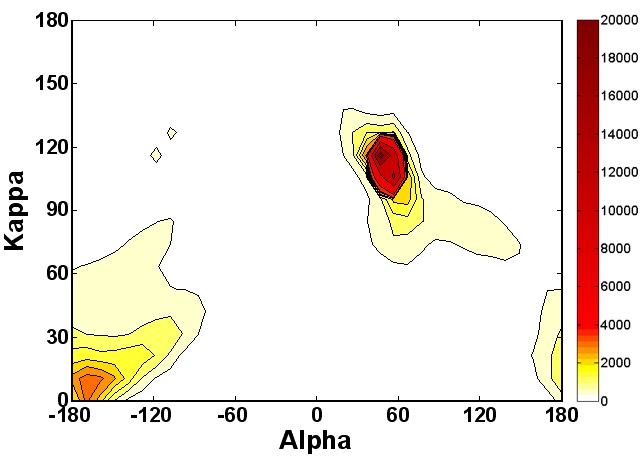
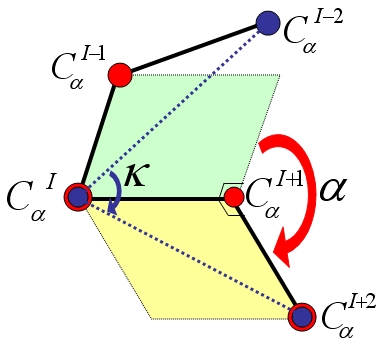
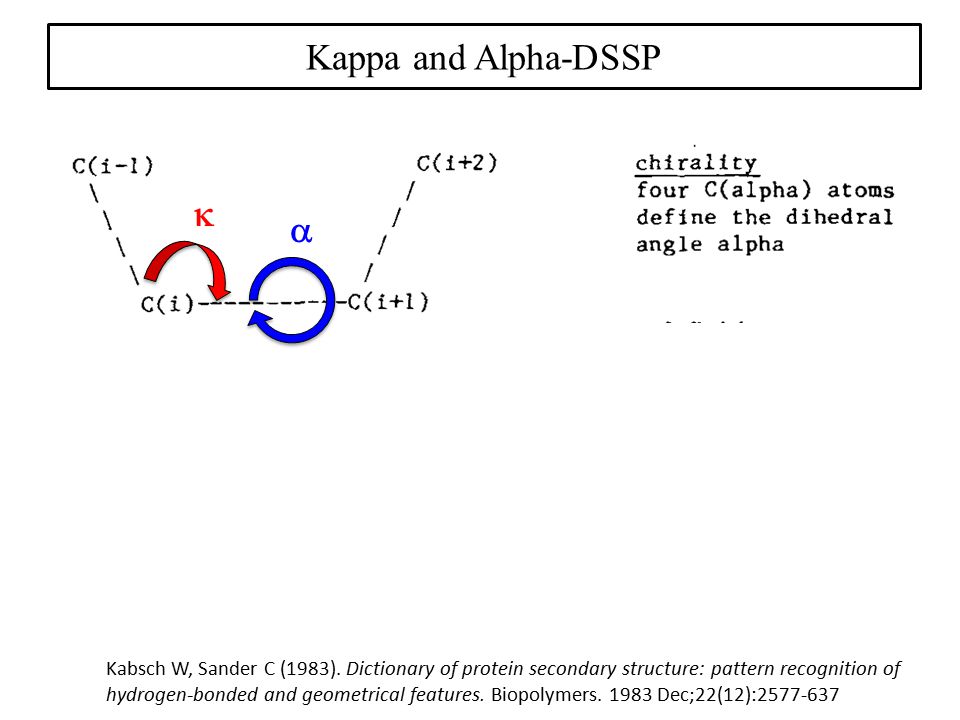
Kappa-alpha plot derived structural alphabet and BLOSUM-like substitution matrix for rapid search of protein structure database | Genome Biology | Full Text
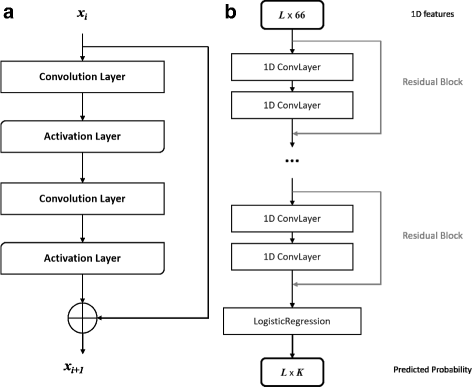
RaptorX-Angle: real-value prediction of protein backbone dihedral angles through a hybrid method of clustering and deep learning | BMC Bioinformatics | Full Text